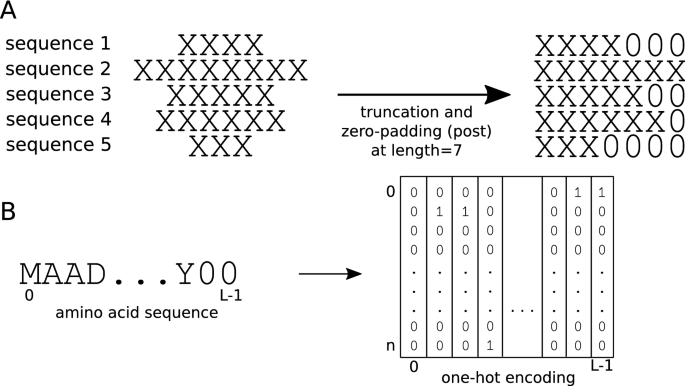
Effect of sequence padding on the performance of deep learning models in archaeal protein functional prediction
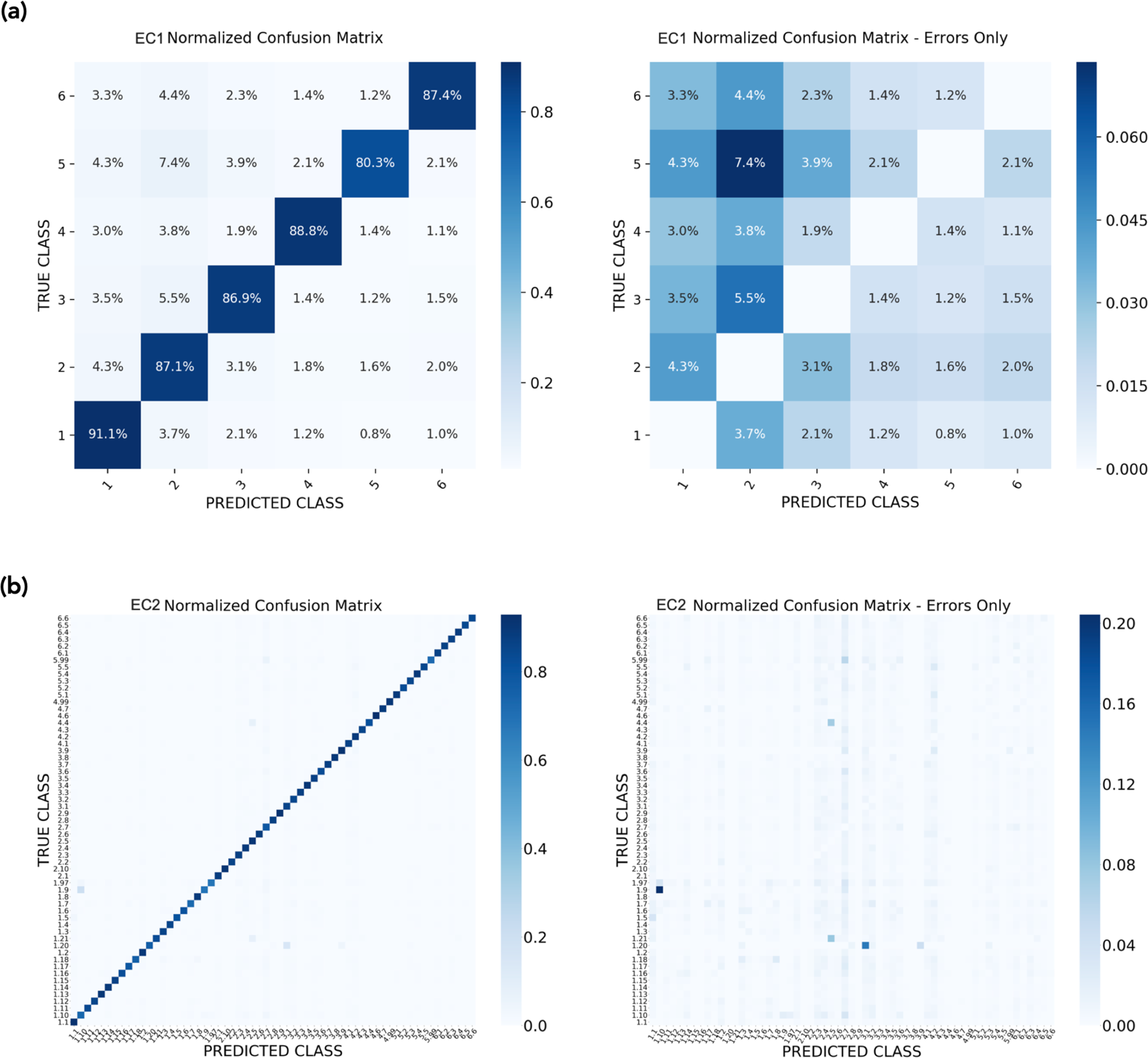
Deep learning of a bacterial and archaeal universal language of life enables transfer learning and illuminates microbial dark matter

MECE: a method for enhancing the catalytic efficiency of glycoside hydrolase based on deep neural networks and molecular evolution - ScienceDirect

From sequence to function through structure: Deep learning for protein design - Computational and Structural Biotechnology Journal

Amino Acid Encoding Methods for Protein Sequences: A Comprehensive Review and Assessment

Discovering molecular features of intrinsically disordered regions by using evolution for contrastive learning

Effect of sequence padding on the performance of deep learning models in archaeal protein functional prediction
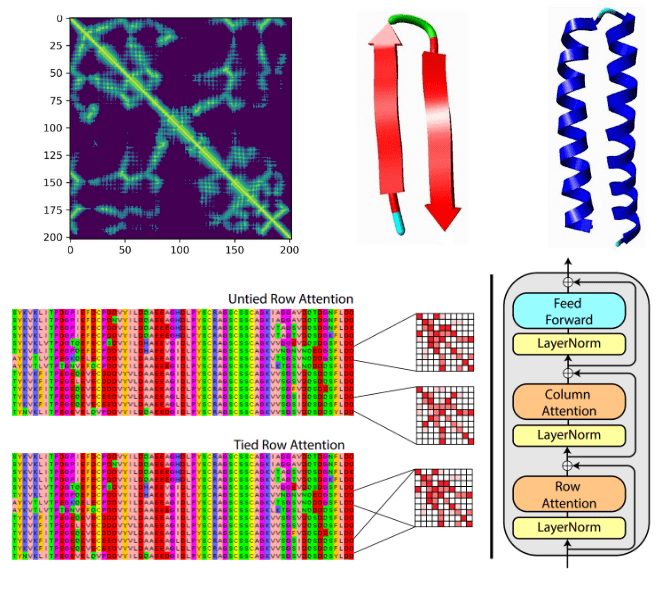
Deep learning on computational biology and bioinformatics tutorial: from DNA to protein folding and alphafold2

FFP: joint Fast Fourier transform and fractal dimension in amino acid property-aware phylogenetic analysis, BMC Bioinformatics

Exploració per tema Aminoàcids