dataset - Is there a way to convert smiles format to TUdataset

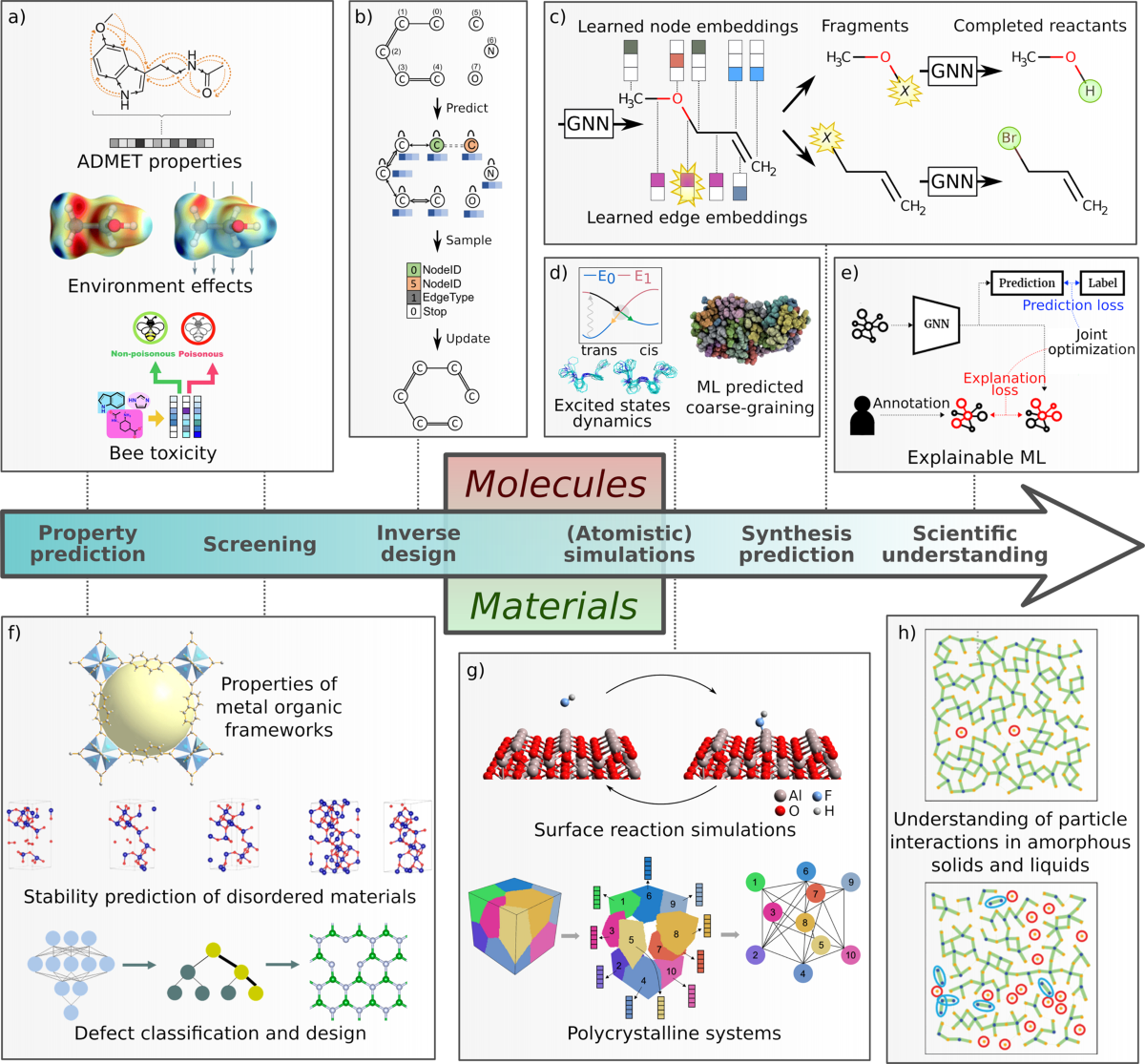
Graph neural networks for materials science and chemistry

PDF) EPIC: Graph Augmentation with Edit Path Interpolation via Learnable Cost

2103.09430] OGB-LSC: A Large-Scale Challenge for Machine Learning on Graphs

PDF) EPIC: Graph Augmentation with Edit Path Interpolation via Learnable Cost

Relating the xyz files to the molecules in PCQM4Mv2 · Issue #297 · snap-stanford/ogb · GitHub

Metapath-fused heterogeneous graph network for molecular property prediction - ScienceDirect
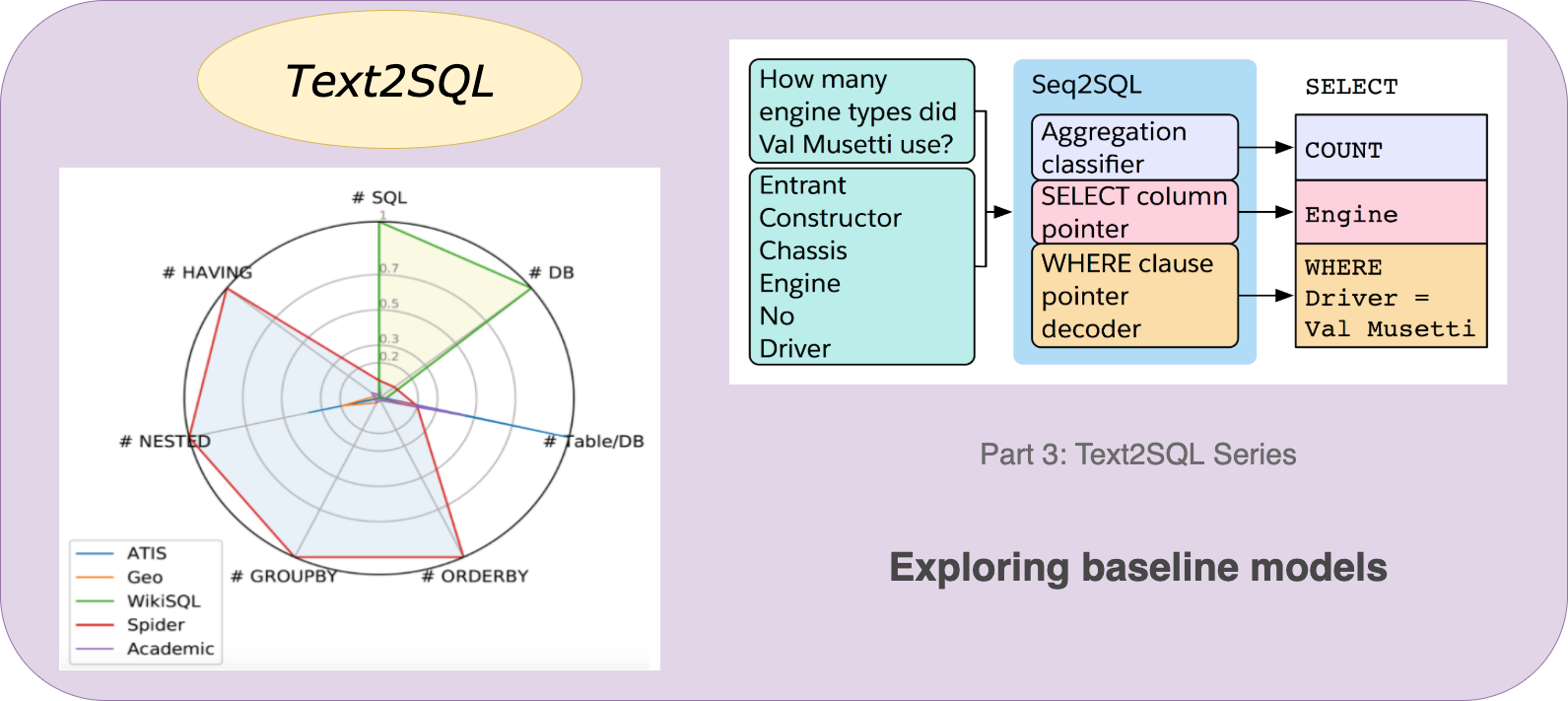
Text2SQL — Part 3: Baseline Models, by Devshree Patel, VisionWizard
GitHub - RohanV01/Molecule_format_converter: This jupyter notebook let's you convert sdf to pdb or SMILES to pdbqt file formats for a batch of compounds to perform cheminformatics and drug discovery projects

Writing a dataset loading script — datasets 1.1.0 documentation

PDF) Visual Analytics For Machine Learning: A Data Perspective Survey

Part 5 - SETAC Europe 22nd Annual Meeting / 6th SETAC World
sdftosmi.py: Convert multiple ligands/compounds in SDF format to SMILES. — Bioinformatics Review