GitHub - tgac-vumc/ACE: Absolute Copy Number Estimation using low-coverage whole genome sequencing data
Absolute Copy Number Estimation using low-coverage whole genome sequencing data - tgac-vumc/ACE

Comprehensive Assessment of Somatic Copy Number Variation Calling Using Next-Generation Sequencing Data

Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data
DNA copy number profiling: from bulk tissue to single cells

Copy number analysis by low coverage whole genome sequencing using ultra low-input DNA from formalin-fixed paraffin embedded tumor tissue, Genome Medicine

GitHub - Nealelab/whole_genome_analysis_pipeline

Accucopy: accurate and fast inference of allele-specific copy number alterations from low-coverage low-purity tumor sequencing data, BMC Bioinformatics
GitHub - DKFZ-ODCF/ACEseqWorkflow: Allele-specific copy number estimation with whole genome sequencing

American Society for Clinical Pharmacology and Therapeutics - 2019 - Clinical Pharmacology & Therapeutics - Wiley Online Library

GATK-gCNV: A Rare Copy Number Variant Discovery Algorithm and Its Application to Exome Sequencing in the UK Biobank

Low-pass whole-genome sequencing in clinical cytogenetics: a validated approach
Bamgineer: Introduction of simulated allele-specific copy number variants into exome and targeted sequence data sets

Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data
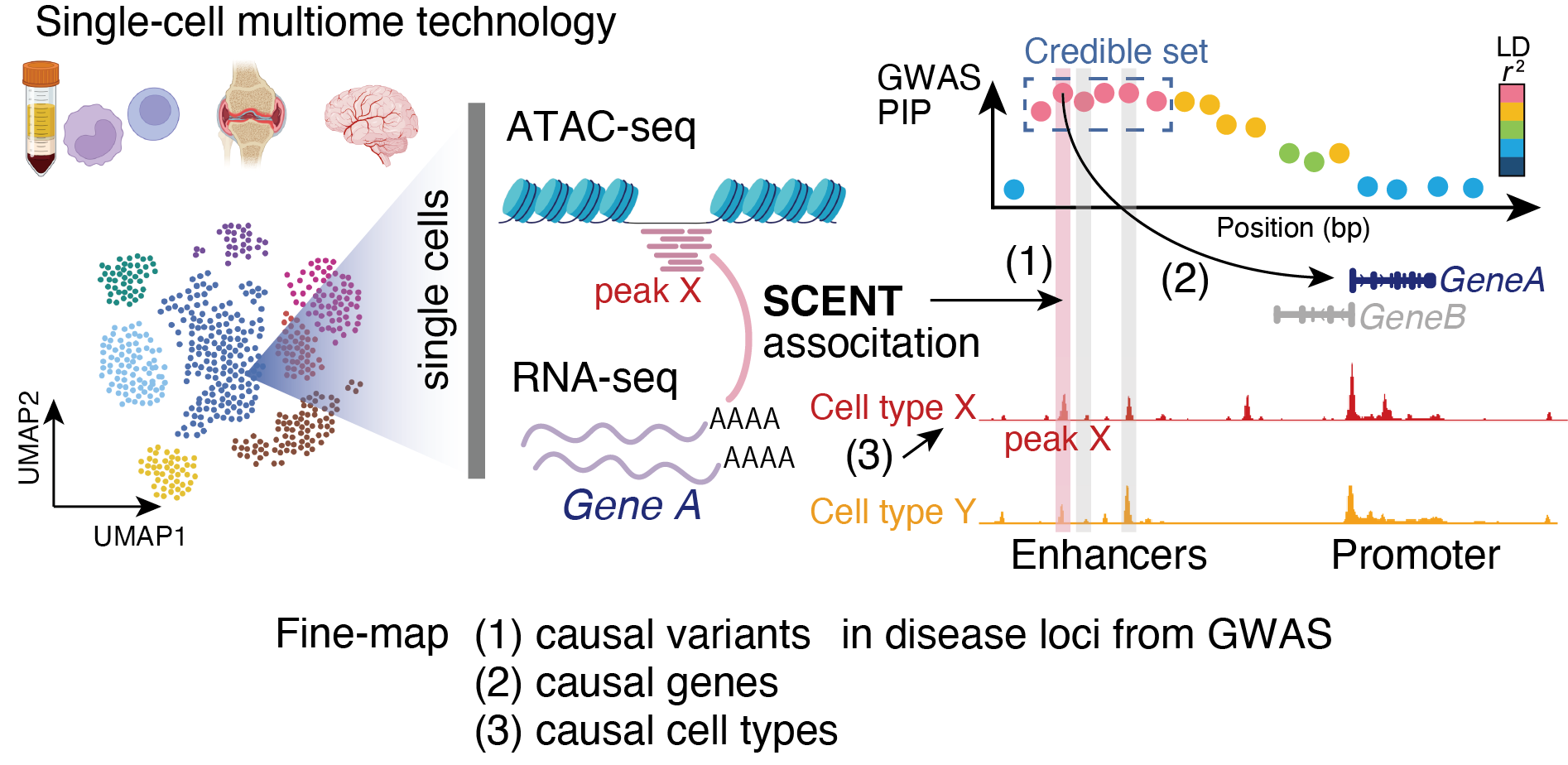
GitHub - immunogenomics/SCENT: Single-Cell ENhancer Target gene mapping using multimodal data with ATAC + RNA

GitHub - Nealelab/whole_genome_analysis_pipeline

Copy-number variants in clinical genome sequencing: deployment and interpretation for rare and undiagnosed disease